How it works#
This page of the documentation is dedicated to explain the theory behind the algorithm, how it is built and present you some key results obtained from experiments conducted on real-world datasets.
First, we’ll detail the problem of clustering under radius constraint, then we’ll explain the Minimum Dominating Set (MDS) problem and how it is adapted to the clustering problem. Finally, we’ll present some key results obtained from experiments conducted on real-world datasets.
Clustering under radius constraint#
Clustering tasks are globally concerned about grouping data points into clusters based on some similarity measure. Clustering under radius constraints is a specific clustering task where the goal is to group data points such that the minimal maximum distance between any two points in the same cluster is less than or equal to a given radius. Mathematically, given a set of data points \(X = \{x_1, x_2, \ldots, x_n\}\) and a radius \(r\), the goal is to find a partition \(\mathcal{P}\) of \(X\) into clusters \(C_1, C_2, \ldots, C_k\) such that : \(\forall C \in \mathcal{P}, \min_{x_i \in C}\max_{x_j \in C}\ d_{ij} \leq r\) where \(d_{ij} = d(x_i, x_j)\) is the dissimilarity between \(x_i\) and \(x_j\).
Minimum Dominating Set (MDS) problem#
The Radius Clustering package implements a clustering algorithm based on the Minimum Dominating Set (MDS) problem. The MDS problem is a well-known NP-Hard problem in graph theory, and it has been proven to be linked to the clustering under radius constraint problem. The MDS problem is defined as follows:
Given a undirected weighted graph \(G = (V,E)\) where \(V\) is a set of vertices and \(E\) is a set of edges, a dominating set \(D\) is a subset of \(V\) such that every vertex in \(V\) is either in \(D\) or adjacent to a vertex in \(D\). The goal is to find a dominating set \(D\) such that the number of vertices in \(D\) is minimized. This problem is known to be NP-Hard.
However, solving this problem in the context of clustering task can be useful. But it has to be adapted to the needs of a clustering task.
Presenting the algorithm#
To adapt the MDS problem to the clustering under radius constraint problem, we need to define a graph based on the data points. The vertices of the graph are the data points, and the edges are defined based on the distance between the data points. The weight of the edges is the dissimilarity between the data points. Then, the algorithm operates as follows:
Construct a graph \(G = (V,E)\) based on the data points \(X\).
Prune the graph by removing the edges \(e_{ij}\) such that \(d(x_i,x_j) > r\).
Solve the MDS problem on the pruned graph.
Assign each vertex to the closest vertex in the dominating set. In case of a tie, assign the vertex to the vertex with the smallest index.
Return the cluster labels.
Experimental results#
The Radius Clustering package provides two algorithms to solve the MDS problem: an exact algorithm and an approximate algorithm. The approximate algorithm [casado] is based on a heuristic that iteratively selects the vertex that dominates the most vertices in the graph. The exact algorithm [jiang] is based on a branch-and-bound algorithm that finds the minimum dominating set in the graph. Experimentation has been conducted on real-world datasets to compare the performances of these two algorithms, and compare them to state-of-the-art clustering algorithms. The complete results from first experiments are available in the paper Clustering under radius constraint using minimum dominating sets.
The algorithms selected for comparison are:
Equiwide clustering (EQW-LP), a state-of-the-art exact algorithm using LP formulation of the problem ([andersen])
Protoclust ([bien]), a state-of-the-art approximate algorithm based on the hierarchical agglomerative clustering using MinMax linkage.
Here are some key results from the experiments:
Dataset |
MDS-APPROX |
MDS-EXACT |
EQW-LP |
PROTOCLUST |
|---|---|---|---|---|
Iris |
3 |
3 |
3 |
4 |
Wine |
4 |
3 |
3 |
4 |
Glass Identification |
7 |
6 |
6 |
7 |
Ionosphere |
2 |
2 |
2 |
5 |
WDBC |
2 |
2 |
2 |
3 |
Synthetic Control |
8 |
6 |
6 |
8 |
Vehicle |
5 |
4 |
4 |
6 |
Yeast |
10 |
10 |
10 |
13 |
Ozone |
3 |
2 |
2 |
3 |
Waveform |
3 |
3 |
3 |
6 |
Dataset |
MDS-APPROX |
MDS-EXACT |
EQW-LP |
PROTOCLUST |
|---|---|---|---|---|
Iris |
1.43 |
1.43 |
1.43 |
1.24 |
Wine |
220.05 |
232.08 |
232.08 |
181.35 |
Glass Identification |
3.94 |
3.94 |
3.94 |
3.31 |
Ionosphere |
4.45 |
5.45 |
5.45 |
5.35 |
WDBC |
1197.42 |
1197.42 |
1197.42 |
907.10 |
Synthetic Control |
66.59 |
70.11 |
70.11 |
68.27 |
Vehicle |
150.87 |
155.05 |
155.05 |
120.97 |
Yeast |
0.42 |
0.42 |
0.42 |
0.42 |
Ozone |
235.77 |
245.58 |
245.58 |
194.89 |
Waveform |
10.73 |
10.73 |
10.73 |
10.47 |
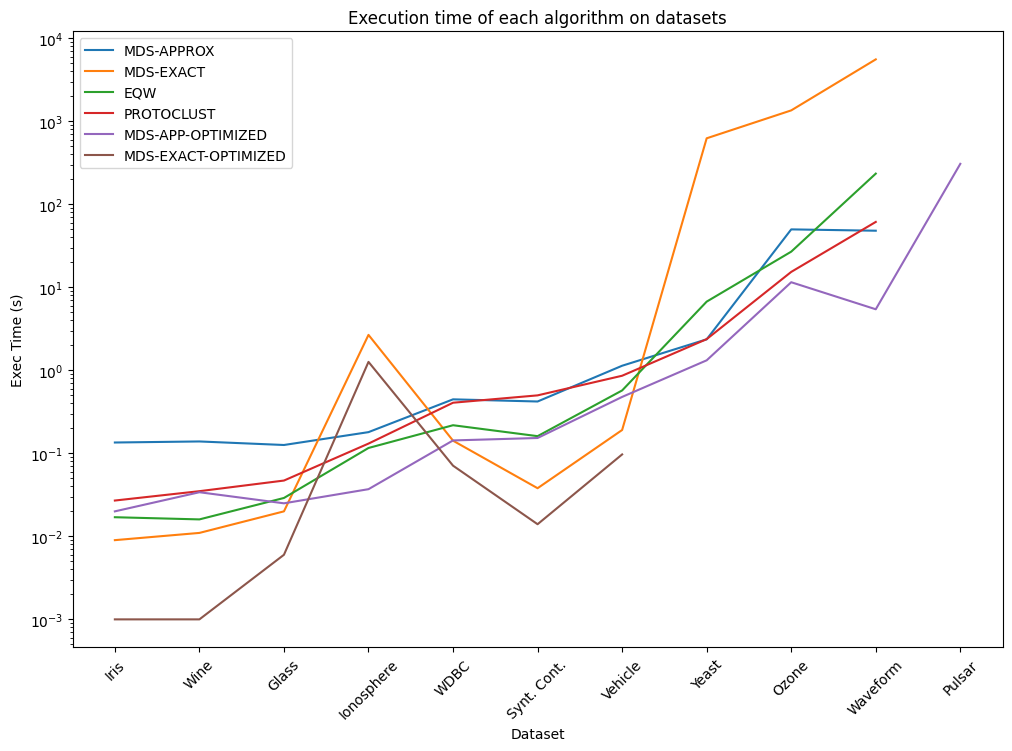
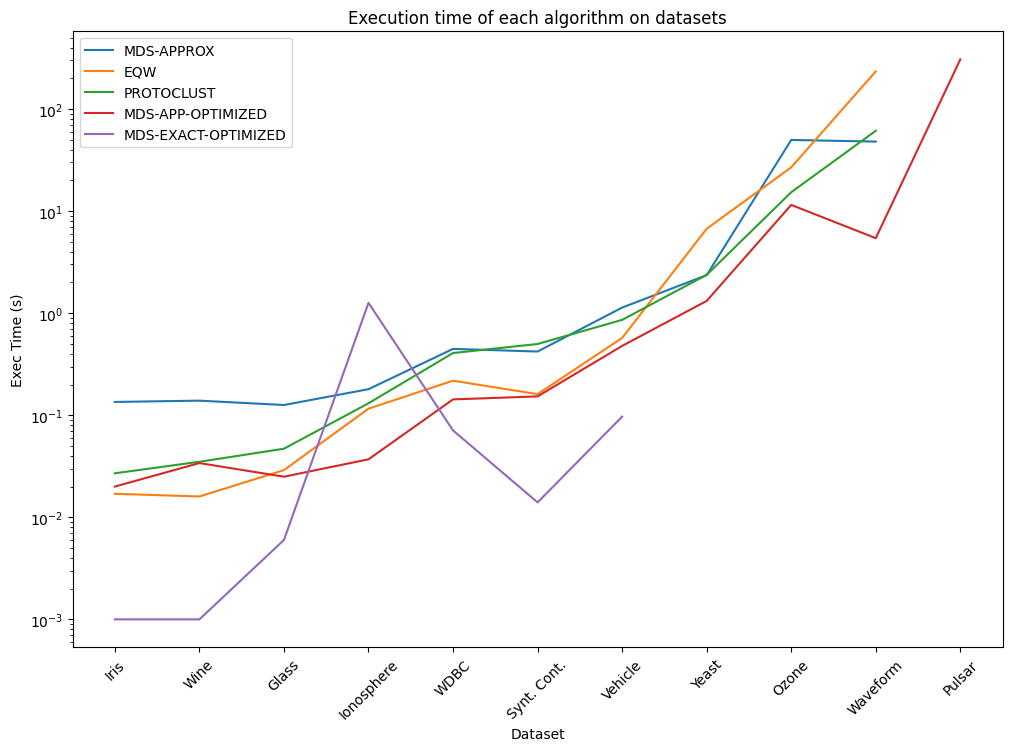
Key insights:#
The approximate algorithm is significantly faster than the exact algorithm, but it may not always provide the optimal solution.
The exact algorithm is slower but provides the optimal solution. Does not scale well to large datasets, due to the NP-Hard nature of the problem.
The approximate algorithm is a good trade-off between speed and accuracy for most datasets.
MDS based approach are both more accurate than Protoclust. However, Protoclust is remarkably faster on most datasets.
Note
The results show that MDS-based clustering algorithms might be a good alternative to state-of-the-art clustering algorithms for clustering under radius constraint problems.
Note
Since the publication of the paper, the Radius Clustering package has been improved and optimized. The results presented here are based on the initial version of the package. For the latest results, please refer to the documentation or the source code.
